Abstract
Clinical outcome of patients with acute myeloid leukemia (AML) is associated with demographic and genetic features. Although the associations of acquired genetic alterations with patients’ sex have been recently analyzed, their impact on outcome of female and male patients has not yet been comprehensively assessed. We performed mutational profiling, cytogenetic and outcome analyses in 1726 adults with AML (749 female and 977 male) treated on frontline Alliance for Clinical Trials in Oncology protocols. A validation cohort comprised 465 women and 489 men treated on frontline protocols of the German AML Cooperative Group. Compared with men, women more often had normal karyotype, FLT3-ITD, DNMT3A, NPM1 and WT1 mutations and less often complex karyotype, ASXL1, SRSF2, U2AF1, RUNX1, or KIT mutations. More women were in the 2022 European LeukemiaNet intermediate-risk group and more men in adverse-risk group. We found sex differences in co-occurring mutation patterns and prognostic impact of select genetic alterations. The mutation-associated splicing events and gene-expression profiles also differed between sexes. In patients aged <60 years>SF3B1 mutations were male-specific adverse outcome prognosticators. We conclude that sex differences in AML-associated genetic alterations and mutation-specific differential splicing events highlight the importance of patients’ sex in analyses of AML biology and prognostication.
Introduction
Acute myeloid leukemia (AML) is a biologically and clinically heterogenous disease that affects a diverse patient population composed of both sexes and all ages [1, 2]. Several pretreatment factors, both disease- and patient-specific, affect prognosis of AML patients [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]. The former include recurrent cytogenetic findings [3,4,5,6,7,8,9,10,11] and gene mutations [12,13,14,15,16,17,18,19] at diagnosis, whereas increasing age is a well-known patient-specific predictor of worse survival [1, 2, 9, 20]. The incidence of acquired cytogenetic and molecular alterations and their prognostic impact vary by age [9] and racial-ethnic identity [21, 22]. Moreover, AML is more common in men than women [1, 23], but the reasons for this sex bias remain largely unknown. Despite the recognition of differences in frequencies of genetic alterations in male and female patients with AML [23, 24], to our knowledge, no large study has yet comprehensively assessed potential associations between patient sex and prognostic impact of recurrent mutations. Therefore, we assessed the frequencies of pretreatment cytogenetic and molecular features and their impact on outcomes of male and female patients in a cohort of 1726 adults with AML in the USA and in a large validation cohort of 954 patients in Germany, both treated on multicenter studies.
Methods
Patients and treatment
We analyzed 1726 adults with de novo AML, including 749 self-reported women and 977 men, who were treated on frontline Cancer and Leukemia Group B (CALGB)/Alliance for Clinical Trials in Oncology (Alliance) protocols. We limited our study to patients with de novo AML in order to avoid the confounding effects of AML type, that is, de novo AML versus AML evolving from an antecedent myelodysplastic syndrome versus therapy-related AML. Self-reported patient sex was confirmed by the results of centrally reviewed [25] metaphase karyotyping. Almost all patients received intensive cytarabine/anthracycline-based frontline treatment on CALGB/Alliance trials between 1986 and 2015 (details are provided in the Supplementary Information). Per study protocols, no patient received an allogeneic hematopoietic stem-cell transplantation (HSCT) in first complete remission (CR), and patients who received an off-protocol HSCT were excluded from analyses of disease-free (DFS) and overall survival (OS) because their follow-up data were either missing or incomplete. Because of differences in the treatment protocols between patients aged <60 years and those aged 60="60" older>Supplementary Information), we performed outcome analyses separately for these age groups. Institutional Review Board approval of all CALGB/Alliance and German AML Cooperative Group (AMLCG) protocols was obtained before any research was performed. Patients provided study-specific written informed consent to participate in treatment studies. Treatment protocols were in accordance with the Declaration of Helsinki. The AMLCG validation cohort of 954 patients (465 female and 489 male) is described in the Supplementary Information.
Mutational profiling
The mutational status of 80 protein-coding genes was determined centrally at The Ohio State University by targeted amplicon sequencing using the MiSeq platform (Illumina, San Diego, CA) [26]. Testing for FLT3 internal tandem duplications (FLT3-ITD) was performed with the Sanger sequencing method [27]. Determination of CEBPAbZIP status was done following 2022 European LeukemiaNet (ELN) guidelines [1] using Miseq panel and/or transcriptional profiling [28]. SF3B1 and SRSF2 were considered as mutated if they concurred with the biology-associated differential splicing events (see details below). Experimental details are provided in the Supplementary Information. Preparation of samples and sequencing in the AMLCG validation cohort followed a comparable workflow [18]. Methods used to analyze gene expression and differential splicing are described in the Supplementary Information.
Clinical endpoints and statistical analysis
Definitions of clinical endpoints—CR, early death, DFS and OS—are provided in the Supplementary Information [29]. Pretreatment features of female and male patients were compared using the Fisher’s exact for categorical variables and Wilcoxon rank-sum tests for continuous variables. Estimated probabilities of DFS and OS were calculated using the Kaplan–Meier method, and the log-rank test evaluated differences between survival distributions [30]. We used logistic regression for modeling CR and Cox proportional hazard regression for interaction and modeling DFS and OS for univariable and multivariable outcome analyses, which were calculated using a limited backward selection technique, and adjusted P values to control for per family error rate. All analyses were performed by the Alliance Statistics and Data Center on a database locked on June 6, 2021, using SAS 9.4, TIBCO Spotfire S+ 8.2 and GraphPad Prism 9.
Results
Clinical and molecular characteristics of AML patients with respect to sex
Our analysis of pretreatment characteristics in the CALGB/Alliance cohort revealed that female patients tended to be younger (median age, 51 vs 54 years, P = 0.06), and had higher white blood cell counts (median, 26.0 vs 22.2 × 109/L, P = 0.009) and percentages of bone marrow blasts (median, 68% vs 65%, P = 0.04). Females had also more often cytogenetically normal AML (CN-AML; 52% vs 42%, P < 0.001), and less often complex karyotype (8% vs 12%, P = 0.005; Supplementary Table 1). Mutational analysis revealed that female patients harbored more often DNMT3A (P < 0.001), NPM1 (P < 0.001) and WT1 (P = 0.02) mutations as well as FLT3-ITD (P = 0.03), and less often ASXL1 (P < 0.001), SRSF2 (P < 0.001), U2AF1 (P = 0.001), RUNX1 (P = 0.04), or KIT (P = 0.05) mutations (Table 1). Notably, all aforementioned genes are located in autosomal chromosomes. We also observed sex-associated differences in the frequencies of mutations in genes categorized into the major AML-associated functional groups, that included female patients having a higher frequency of mutations in methylation-related genes (51% vs 46%, P = 0.02) and male patients having a higher frequency of mutations in genes involving chromatin remodeling (23% vs 18%, P = 0.02), spliceosome (22% vs 11%, P < 0.001), and, by trend, transcription factors (28% vs 24%, P = 0.06); men also harbored myelodysplasia-related gene mutations more often than women (37% vs 27%, P < 0.001; Table 2).
To validate the observed sex-associated differences, we compared the frequencies of cytogenetic findings (Supplementary Table 1) and gene mutations in patients from AMLCG (Tables 1 and 2). The results were largely concordant, with CN-AML and all mutations, except KIT and WT1 mutations, which differed between males and females among CALGB/Alliance patients being also significantly different in the AMLCG cohort. However, in the latter patient population, females also less often carried EZH2 (P = 0.005), SMC1A (P = 0.003) and STAG2 (P < 0.001) mutations and mutations in the cohesion complex genes (P < 0.001) than males.
The proportions of patients assigned to genetic-risk groups in the 2022 ELN classification [1] also differed between sexes. In the CALGB/Alliance cohort, a higher percentage of female patients was categorized in the intermediate-risk group (30% vs 20%) and a lower percentage of women was included in the adverse-risk group (31% vs 44%, Supplementary Table 1).
Sex-associated differences in co-occurring molecular alterations
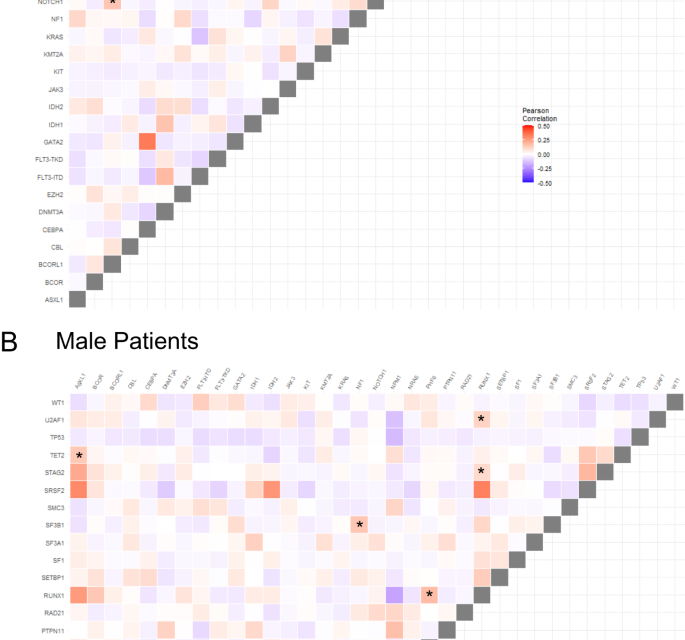
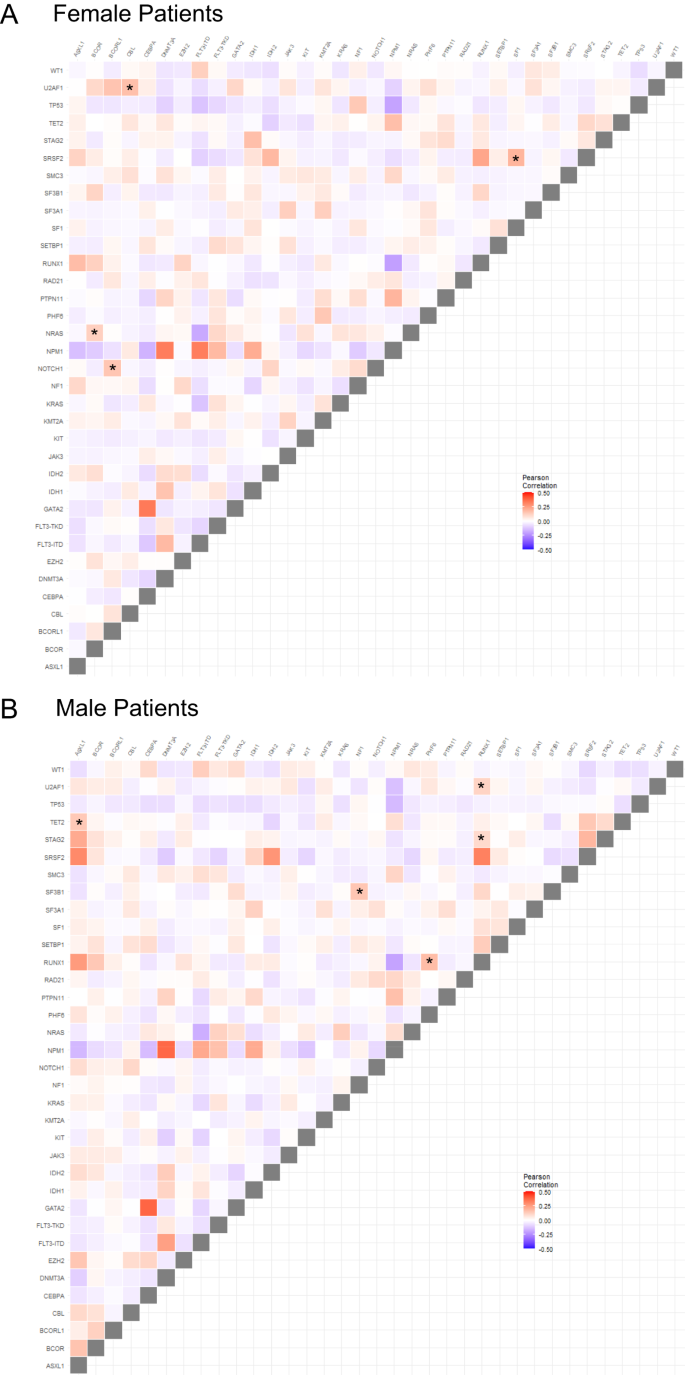
We next compared patterns of co-occurring recurrent mutations in male and female patients and differences between sexes (Pearson correlation <0>P < 0.0002 for one sex and Pearson correlation >0.03 and P > 0.5). We identified four gene mutation pairs, including NRAS/BCOR, U2AF1/CBL, NOTCH1/BCORL1, and SF1/SRSF2 whose presence was strongly associated with female patients. Conversely, ASXL1/TET2, NF1/SF3B1, RUNX1/PHF6, RUNX1/U2AF1 and RUNX1/STAG2 were strongly positively associated with male patients. Conversely, WT1/SRSF2 tended to be mutually exclusive in males but less so in females (Fig. 1).
Treatment outcome of adults with AML aged <60 years with respect to patient sex>
We analyzed clinical outcomes of 381 female and 463 male AML patients aged 17–59 years in the CALGB/Alliance cohort, for whom frontline treatment with intensive induction chemotherapy is currently still a standard of care. We found no significant differences in CR and early death rates, DFS or OS between female and male AML patients (Supplementary Table 2). There were also no differences between sexes in CR and early death rates or DFS in the AMLCG cohort. However, male German patients aged 18–59 years had a shorter OS than female patients (5-year rates, 42% vs 51%, P = 0.005).
Sex-associated prognostic impact of genetic alterations in patients aged < 60 years
Since cytogenetic and molecular genetic findings are routinely used for risk stratification of AML patients, we assessed sex-specific outcomes based on the 2022 ELN genetic-risk categories [1]. Despite differences in proportions of females and males in the intermediate- and adverse-risk groups, DFS and OS was essentially equal within each genetic-risk group, except for longer OS of female adverse-risk patients (P < 0.001; Supplementary Fig. 1A, B).
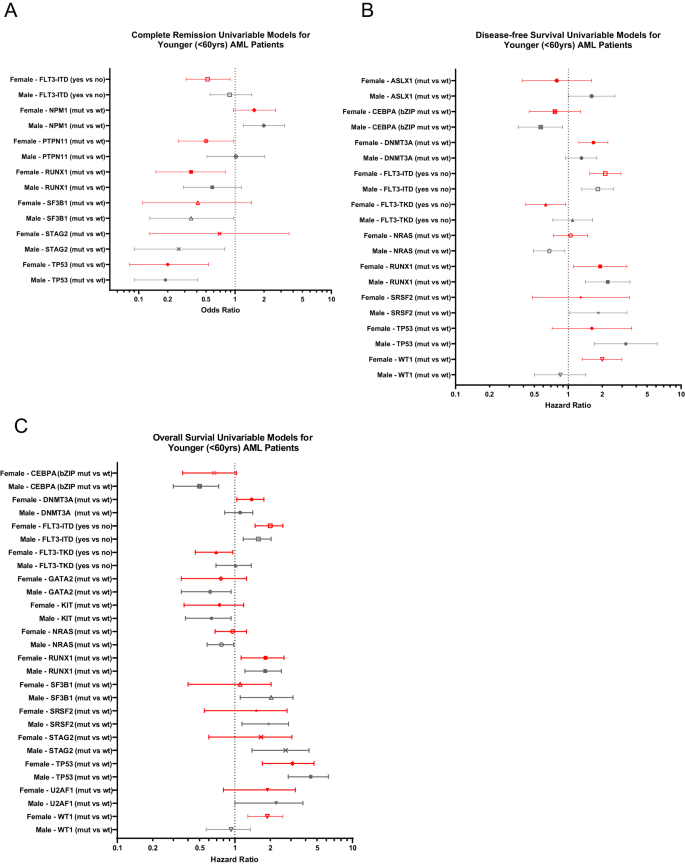
Next, we assessed associations of recurrent genetic alterations with outcome of CALGB/Alliance patients. In univariable analysis (UVA) for CR, presence of FLT3-ITD, PTPN11 and RUNX1 mutations affected outcome of only female patients (Fig. 2A), of which FLT3-ITD and PTPN11 mutations remained significantly associated with CR achievement also in subsequent multivariable analysis (MVA, Supplementary Table 3). In contrast, a normal karyotype and SF3B1 mutations were adverse predictors only in male patients in both UVA and MVA.
Forest plot illustrating univariable analyses of A complete remission, B disease-free survival and C overall survival. Depicted in the plot are all gene mutations with sex-specific survival impact for either male or female patients in univariable analyses for the respective outcome endpoint. Results of the corresponding multivariable analyses and markers with significance are shown in Supplementary Table 3.
For DFS in female patients, DNMT3A and WT1 mutations associated with shorter, and FLT3-TKD mutations with longer DFS in both UVA and MVA (Fig. 2B, Supplementary Fig. 2A). While for male patients detection of ASXL1, CEBPAbZIP, NRAS, SRSF2 and TP53 mutations had sex-specific survival impact in UVA, only CEBPAbZIP mutations were significant in multivariable models.
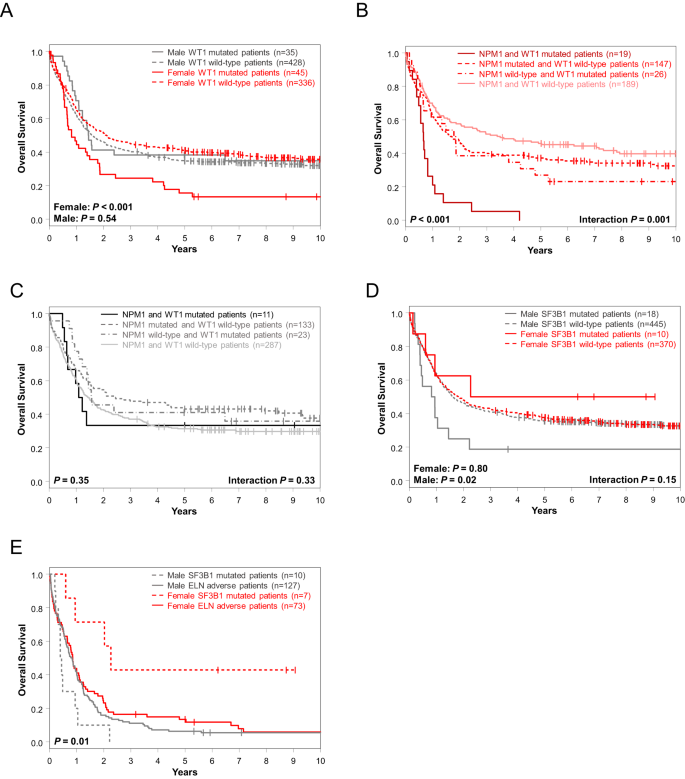
In univariable analysis of OS, FLT3-TKD, DNMT3A and WT1 mutations affected outcome of only female patients (Fig. 2C) of which, again, WT1 mutations held their significance also in multivariable analyses (Fig. 3A). Notably, the adverse outcome impact of female WT1-mutated patients seemed to be driven by WT1/NPM1 co-mutated female patients (Fig. 3B, C). OS of men only was influenced by CEBPAbZIP, GATA2, KIT, NRAS, SF3B1, SRSF2, STAG2 and U2AF1 mutations in UVA, of which SF3B1 mutations had again sex-specific survival association in MVA, too (Fig. 3D, Supplementary Table 3).
A Overall survival of female and male patients with and without WT1 mutations and overall survival of B female and C male patients according to their NPM1 and WT1 mutation status. D Overall survival of female and male patients with and without SF3B1 mutations. E Overall survival of female and male patients classified in the 2022 ELN adverse-risk group because of the presence of SF3B1 mutation (and lack of favorable-risk genetic markers) and of the remaining female and male patients classified in the 2022 ELN adverse-risk group who did not have SF3B1 mutation.
Focusing on those sex-specific molecular alterations that held their significance in both UV and MV models and at least 2 survival endpoints (female: WT1 mutations [DFS, OS]; male: SF3B1 mutations [CR, OS]), we tested their outcome impacts in the AMLCG cohort.
While WT1 mutations associated with a trend for inferior DFS in female AMLCG patients, there was no association with inferior OS, possibly suggestive of a rescue effect of more frequently used intensive consolidation including allogeneic HSCT which is more commonly administered in Germany (Supplementary Fig. 2B, C). Although limited by relatively small sample sizes, SF3B1-mutated male patients in the AMLCG cohort also had a lower CR rate (38% vs 72%, P = 0.04) and tended to have inferior OS (Supplementary Fig. 3).
As SF3B1 mutations demonstrated distinct prognostic differences and were recently added to the 2022 ELN classification as adverse-risk outcome prognosticator (in the absence of favorable-risk genetic markers), we compared OS of adverse-risk female and male patients with SF3B1 mutations and OS of the remaining adverse-risk female and male patients (i.e., without SF3B1 mutations). While male SF3B1-mutated 2022 ELN adverse-risk patients and both female and male adverse-risk patients without SF3B1 mutations had similarly poor overall survival, female SF3B1-mutated adverse-risk patients had significantly longer OS (5-year rates, 43% vs 8%, P = 0.01) than other 2022 ELN adverse-risk patients (Fig. 3E).
Treatment outcome of adults with AML aged ≥60 years with respect to patient sex and the sex-associated prognostic impact of genetic alterations
Given the generally poor survival of older patients with AML, we performed a subset outcome analyses of the AML patients aged 60 years and older included in our study. We did not find significant differences between sexes among 524 older patients treated on the CALGB/Alliance protocols nor among 414 older German patients (Supplementary Table 4). There were no significant differences in DFS or OS between female and male patients in any of the 2022 ELN genetic-risk groups, with patients in the intermediate-risk and adverse-risk groups performing similarly poorly (Supplementary Fig. 4A, B). Although drawing definitive conclusions from the multivariable analyses is difficult because of the generally poor treatment outcome of older patients with AML, we found that such mutations included in the 2022 ELN genetic-risk classification as FLT3-ITD, NPM1 and TP53 mutations were among main factors affecting CR rates and survival (Supplementary Table 5). Only in men, DFS was also negatively affected by PTPN11 mutations and OS by KRAS mutations.
Sex-specific alternative splicing events and lineage-associated gene expression patterns
To investigate the potential role of sex bias on gene and splicing programs, we next analyzed patients with existing RNAseq data (n = 848) to quantify alternative splicing events (ASE) and differentially expressed genes (DEGs) associated with recurrent gene mutations or cytogenetic findings using the AltAnalyze workflow, including those affecting spliceosome genes and other recurrent AML-associated genes or cytogenetic features (n = 24) and select clinical/patient demographic variables.
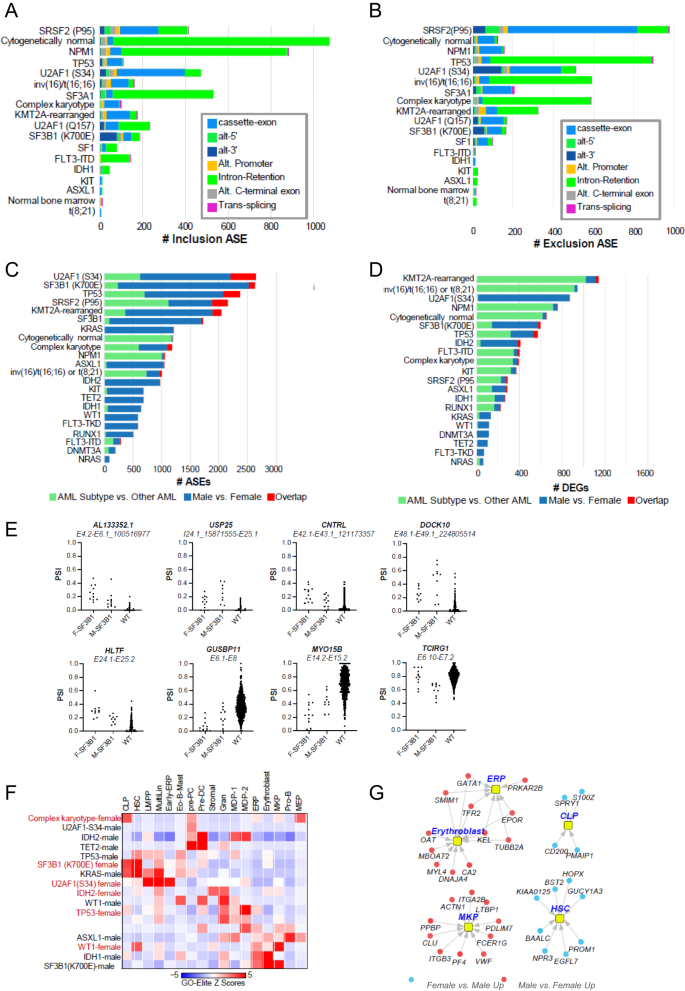
To set the stage, we first determined the spectrum of mutations, which result in common splicing impacts via supervised and unsupervised analyses irrespective of sex, which could be verified in independent AML cohorts (Supplementary Methods and Supplementary Fig. 5A, B). Most AML-associated mutations and chromosome rearrangements resulted in splicing impacts, most significantly associated with splicing-factor mutations (SRSF2-P95*, U2AF1-S34*, SF3B1, SF3A1, U2AF1-Q157*, SF1), common mutations (NPM1, TP53), CN-AML, complex karyotype, and chromosome rearrangements resulting in gene fusions involving CBFB and KMT2A (Fig. 4A, B).
The extent of exon/intron A inclusion or B exclusion ASEs. The extent of unique sex-associated ASEs C or differentially expressed genes (DEGs) D associated with each patient subtype, unique regulated events or genes compared with all other AMLs and sex/subtype overlapping events/genes, derived from the software AltAnalyze. Green color denotes the number of unique ASEs or DEGs in all patients with the indicated molecular genetic or cytogenetic subtype versus all other AML patients in the cohort. Blue indicates the number of unique ASEs or DEGs in males versus females in the indicated AML subtype and red illustrates overlap between subtype-associated ASEs or DEGs and male versus female (subtype regulated events/genes that vary according to patient sex). E Quantification of SF3B1-mutated sex-associated alternative splicing events as percent spliced in (PSI) values for the indicated parental exon-exon junction. F = female, M = Male. F Heatmap of gene set enrichments (GO-Elite) for known AML subtypes against human bone marrow cell-type markers to identify lineage skewing, BM cell-type markers were derived from extensive prior human single-cell analyses [43], to identify markers of rare hemopoietic stem cells, progenitors, and immune cell-types. Only AML subtype sex-associated gene sets with significant enrichments are shown. G Sex-associated differentially expressed genes in SF3B1-mutated males versus females corresponding to human bone marrow progenitor populations.
Next, we compared female and male patients in each of these defined subtypes. We considered subtypes with >17 samples and an unadjusted P-value for these comparisons. Interestingly, we found different magnitudes of splicing and gene-expression impacts associated with sex in AML subtypes, with generally greater impacts associated with ASEs versus DEGs (Fig. 4C, D). In general, splicing-factor mutations (SRSF2-P95*, U2AF1-S34*, SF3B1, SF3A1, SF3A1, SF1) resulted in large sex-associated differences for both ASEs and DEGs. Notably, while it has been extensively documented that SF3B1-K700E and functionally similar mutations result in alternative splice sites only selected for within the presence of these mutations, examination of SF3B1-K700E and related mutation ASEs predicted sex bias for several of previously annotated (USP25, MYO15B) and novel SF3B1-K700E selected cryptic splice sites (AL133352.1, CNTRL, DOCK10), in addition to non-cryptic splice sites with clear sex bias (Fig. 4E). At the pathway level, we found that SF3B1 mutations in male patients transcriptionally induce genes associated with integrin cell-surface receptor-linked signaling pathways (MAP kinase, Netrin, PECAM1, Ephrin B, interferon alpha/beta signaling, L1CAM), whereas RUNX1- and TP53-mutated male patients induce inflammatory response pathways (IL12, FGF, CXCR4 and TCR signaling). Further, complex karyotype and TP53 mutations in female patients induce a distinct set of inflammatory and immune signaling programs (Supplementary Fig. 5C, D). When we considered marker genes for hematopoietic human progenitor and differentiated cell populations, we noted distinct lineage enrichments shared by different sets of mutations. In particular, SF3B1 (females) and KRAS (males) were enriched in hematopoietic stem cells, MultiLin, common lymphoid precursor and eosinophil/mast programs, whereas SF3B1 (males) and WT1 (females) induced a dominant megakaryocytic progenitors/erythroid progenitors (Fig. 4F). We note that in male SF3B1-mutated patients, genes associated with these programs represent well-defined lineage determining factors, such as HOPX and PROM1 for hematopoietic stem cells, ITGA2B, VWF and PF4 for megakaryocytic progenitors and EPOR and GATA1 for erythroid progenitors (Fig. 4G). Together, these data indicate that sex contributes to both alternative splicing and transcriptional differences among patients with splicing factor and other dominant AML mutations.
Discussion
The discovery of recurrent cytogenetic and molecular genetic alterations has improved our understanding of AML biology and resulted in the routine use of pretreatment genetic alterations for risk stratification [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]. However, genetic changes should not be assessed in isolation, because their effects may be influenced by such factors as patients age [1, 2, 9, 20] and/or racial-ethnic identity [21, 22]. Whereas female and male patients with AML share the vast majority of genetic information, except for at least 26 protein-coding genes located on the Y chromosome, large-scale genomic studies of health and disease have revealed profound differences in sex-biased gene-regulatory networks [31] and splicing events [32] contributing to phenotypic sex differences.
Recently, De-Morgan et al. [23] also demonstrated specific preleukemic genes that were more frequently mutated in men than women with AML. Consistent with previous reports, our study found a female predominance of such most common (de novo) AML-associated mutations as NPM1, DNMT3A and FLT3-ITD, and a male predominance of spliceosome complex gene mutations and other myelodysplasia-related genes, many of which are found on the X-chromosome (BCOR, BCORL1, SMC1A, STAG2, ZRSR2). These findings might be interesting in the context of the known higher incidence of AML in males compared to females, and suggestive of sex-specific differences in the disease biology. Interestingly, analyzing gene expression and alternative splicing patterns first on a global scale followed by sex-specific analyses, we identified differential splicing events that associated with sex, as well as cell lineage, depending on the gene mutation. Surprisingly, our data suggest that shared pathway-level impacts for pairs of mutations exist, which tend to be associated with sex, whereas cell-type associated impacts indicate a lineage bias in programs, which often differ from the pathway. Although biologic reasons for the male-specific outcome association of SF3B1 mutations are still unclear, the differential effects on cell lineages suggested by GSEA indicate that sex-specific cell programs influencing leukemogenesis and leukemic cell responses may exist.
We detected male-specific association of SF3B1 mutations with higher resistance to induction chemotherapy and shorter overall survival. Notably, SF3B1 mutations, in the absence of favorable-risk genetic markers, have recently been added as an adverse-risk prognosticator to the 2022 ELN genetic-risk classification [1]. Our data expand upon the work of the GenoMed4All consortium and Maggioni et al. [33], which also demonstrated sex-specific biases at the single-gene level in myelodysplastic syndromes (MDS) and a male specificity for co-mutational pathways in splicing-related genes. Our work, if confirmed, ultimately suggest that SF3B1 mutations might constitute a sex-specific prognosticator, which could be taken into account in the future revisions of the ELN genetic-risk classification.
The negative prognostic impact of WT1 mutations has been repeatedly reported in the past [34,35,36,37,38,39], but their association with female sex have hitherto not been recognized. In our study, an association of WT1 mutations with women was seen only in the CALGB/Alliance cohort, but was not observed in the AMLCG patients. As patients treated in Europe receive allogeneic HSCT in first CR more often, it is possible that more intensive consolidation might alleviate the adverse prognostic impact of WT1 mutations. This discrepancy might warrant further investigation in prospective studies, to help determine the appropriate consolidation strategy for female patients carrying WT1 mutations. Moreover, adverse outcome of WT1-mutated women seems to be associated with co-existence of NPM1 mutations, which was previously suggested to confer high treatment resistance and poor survival rates [17, 40].
Limitations of our study include the fact that we studied only patients with de novo AML, and that the Alliance patients did not undergo allogeneic HSCT in first CR and were almost exclusively treated with intensive induction chemotherapy followed by consolidation therapy. Hence, future studies are needed to assess associations between sex and molecular features and survival in patients undergoing allogeneic HSCT and in those receiving emerging targeted therapies, which is particularly important in older patients. Furthermore, we detected sex-associated differences in the frequencies of myelodysplasia-related gene mutations among patients diagnosed with de novo AML, not in patients with AML evolving from an antecedent MDS whom we did not analyze. It will be thus important to perform dedicated analyses in patients with secondary AML evolving from MDS, especially since sex disparities with respect to genotypes, phenotypes and outcomes have previously been reported in patients diagnosed with MDS [41, 42].
In summary, our study assessed potential sex-associated biologic and prognostic implications of genetic alterations in large patient cohorts. Although most established genetic alterations affected outcomes of both sexes, select gene mutations were associated with patient sex. This was likely missed previously because of routinely performed sex-pooled outcome analyses. We identified SF3B1 mutations as a male-specific and WT1 mutations as female-specific prognosticators of poor survival in patients treated with conventional chemotherapy. Differences in the frequencies of AML-associated genetic alterations and mutation-specific differential splicing events, with possible subsequent phenotypic changes, highlight the importance of considering the patients’ sex in analyses examining leukemia biology and prognostic significance of genetic alterations and their role in clinical decision-making in AML.
Data availability
Patient data used in survival analyses were obtained from the Alliance Statistics and Data Management Center. Individual participant data will not be shared.
References
-
Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77.
-
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52.
-
Byrd JC, Mrózek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461). Blood. 2002;100:4325–36.
-
Arthur DC, Berger R, Golomb HM, Swansbury GJ, Reeves BR, Alimena G, et al. The clinical significance of karyotype in acute myelogenous leukemia. Cancer Genet Cytogenet. 1989;40:203–16.
-
Visani G, Bernasconi P, Boni M, Castoldi GL, Ciolli S, Clavio M, et al. The prognostic value of cytogenetics is reinforced by the kind of induction/consolidation therapy in influencing the outcome of acute myeloid leukemia – analysis of 848 patients. Leukemia. 2001;15:903–9.
-
Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–36.
-
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354–65.
-
Grimwade D, Mrózek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am. 2011;25:1135–61.
-
Mrózek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30:4515–23.
-
Döhner H, Dolnik A, Tang L, Seymour JF, Minden MD, Stone RM, et al. Cytogenetics and gene mutations influence survival in older patients with acute myeloid leukemia treated with azacitidine or conventional care. Leukemia. 2018;32:2546–57.
-
Mrózek K. Molecular cytogenetics in acute myeloid leukemia in adult patients: practical implications. Pol Arch Intern Med. 2022;132:16300.
-
Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–89.
-
Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74.
-
Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Görlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128:686–98.
-
Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21.
-
Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–31.
-
Eisfeld A-K, Kohlschmidt J, Mims A, Nicolet D, Walker CJ, Blachly JS, et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 years>
-
Herold T, Rothenberg-Thurley M, Grunwald VV, Janke H, Goerlich D, Sauerland MC, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34:3161–72.
-
Duncavage EJ, Schroeder MC, O’Laughlin M, Wilson R, MacMillan S, Bohannon A, et al. Genome sequencing as an alternative to cytogenetic analysis in myeloid cancers. N Engl J Med. 2021;384:924–35.
-
Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, et al. Age and acute myeloid leukemia. Blood. 2006;107:3481–5.
-
Bhatnagar B, Kohlschmidt J, Mrózek K, Zhao Q, Fisher JL, Nicolet D, et al. Poor survival and differential impact of genetic features of black patients with acute myeloid leukemia. Cancer Discov. 2021;11:626–37.
-
Sekeres MA, Peterson B, Dodge RK, Mayer RJ, Moore JO, Lee EJ, et al. Differences in prognostic factors and outcomes in African Americans and whites with acute myeloid leukemia. Blood. 2004;103:4036–42.
-
De-Morgan A, Meggendorfer M, Haferlach C, Shlush L. Male predominance in AML is associated with specific preleukemic mutations. Leukemia. 2021;35:867–70.
-
Hellesøy M, Engen C, Grob T, Löwenberg B, Valk PJM, Gjertsen BT. Sex disparity in acute myeloid leukaemia with FLT3 internal tandem duplication mutations: implications for prognosis. Mol Oncol. 2021;15:2285–99.
-
Mrózek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–44.
-
Eisfeld A-K, Mrózek K, Kohlschmidt J, Nicolet D, Orwick S, Walker CJ, et al. The mutational oncoprint of recurrent cytogenetic abnormalities in adult patients with de novo acute myeloid leukemia. Leukemia. 2017;31:2211–8.
-
Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a Cancer and Leukemia Group B study. Cancer Res. 2001;61:7233–9.
-
Taube F, Georgi JA, Kramer M, Stasik S, Middeke JM, Röllig C, et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: differential impact of bZIP and TAD mutations on outcome. Blood. 2022;139:87–103.
-
Cheson BD, Cassileth PA, Head DR, Schiffer CA, Bennett JM, Bloomfield CD, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–9.
-
Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE. Regression methods in biostatistics: linear, logistic, survival and repeated measure models. New York, NY, USA: Springer; 2005.
-
Lopes-Ramos CM, Chen CY, Kuijjer ML, Paulson JN, Sonawane AR, Fagny M, et al. Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 2020;31:107795.
-
Togami K, Chung SS, Madan V, Booth CAG, Kenyon CM, Cabal-Hierro L, et al. Sex-biased ZRSR2 mutations in myeloid malignancies impair plasmacytoid dendritic cell activation and apoptosis. Cancer Discov. 2021;12:522–41.
-
GenoMed4All consortium. A sex-informed approach to improve the personalised decision making process in myelodysplastic syndromes: a multicentre, observational cohort study. Lancet Haematol. 2023;10:e117–e28.
-
Paschka P, Marcucci G, Ruppert AS, Whitman SP, Mrózek K, Maharry K, et al. Wilms’ tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–602.
-
Virappane P, Gale R, Hills R, Kakkas I, Summers K, Stevens J, et al. Mutation of the Wilms’ tumor 1 gene is a poor prognostic factor associated with chemotherapy resistance in normal karyotype acute myeloid leukemia: the United Kingdom Medical Research Council Adult Leukaemia Working Party. J Clin Oncol. 2008;26:5429–35.
-
Renneville A, Boissel N, Zurawski V, Llopis L, Biggio V, Nibourel O, et al. Wilms tumor 1 gene mutations are associated with a higher risk of recurrence in young adults with acute myeloid leukemia: a study from the Acute Leukemia French Association. Cancer. 2009;115:3719–27.
-
Becker H, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Margeson D, et al. Mutations of the Wilms tumor 1 gene (WT1) in older patients with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116:788–92.
-
Hou H-A, Huang T-C, Lin L-I, Liu C-Y, Chen C-Y, Chou W-C, et al. WT1 mutation in 470 adult patients with acute myeloid leukemia: stability during disease evolution and implication of its incorporation into a survival scoring system. Blood. 2010;115:5222–31.
-
Bhatnagar B, Kohlschmidt J, Orwick SJ, Buelow DR, Fobare S, Oakes CC, et al. Framework of clonal mutations concurrent with WT1 mutations in adults with acute myeloid leukemia: Alliance for Clinical Trials in Oncology study. Blood Adv. 2023;7:4671–5.
-
El Hussein S, DiNardo CD, Takahashi K, Khoury JD, Fang H, Furudate K, et al. Acquired WT1 mutations contribute to relapse of NPM1-mutated acute myeloid leukemia following allogeneic hematopoietic stem cell transplant. Bone Marrow Transplant. 2022;57:370–6.
-
Karantanos T, Gondek LP, Varadhan R, Moliterno AR, DeZern AE, Jones RJ, et al. Gender-related differences in the outcomes and genomic landscape of patients with myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes. Br J Haematol. 2021;193:1142–50.
-
Tinsley-Vance SM, Ali NA, Ball S, Aguirre LE, Jain AG, Hussaini MO, et al. Sex disparities in myelodysplastic syndromes: genotype, phenotype, and outcomes. Clin Lymphoma Myeloma Leuk. 2023;23:355–9.
-
Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol. 2018;68:51–61.
-
Mrózek K, Kohlschmidt J, Blachly JS, Nicolet D, Carroll AJ, Archer KJ, et al. Outcome prediction by the 2022 European LeukemiaNet genetic-risk classification for adults with acute myeloid leukemia: an Alliance study. Leukemia. 2023;37:788–98.
Acknowledgements
This study celebrates the life and accomplishments of Dr. Clara D. Bloomfield (1942–2020), who died unexpectedly on March 1, 2020. The authors are grateful to the patients who consented to participate in these clinical trials and the families who supported them; to Christopher Manring and the CALGB/Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services; and to Lisa J. Sterling for data management. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology), UG1CA233253, UG1CA23318, UG1CA283338, UG1CA189824, UG1CA18985, UG1CA233247, UG1CA233338, U10CA140158, UG1CA233331, U10CA180867, R35CA197734, UG1CA189850, and 5P30CA016058; R01 CA262496 (A-KE, ASM); the Coleman Leukemia Research Foundation; The D Warren Brown Foundation; the Pelotonia Fellowship Program (A-KE), ASH Junior Faculty Scholar and ASH Bridge Award (A-KE); the Leukemia & Lymphoma Society Translational Research Grant (A-KE); The Leukemia Research Foundation (A-KE); The American Cancer Society (A-KE), the National Comprehensive Cancer Network Foundation Young Investigator Award (JSB); the Alliance for Clinical Trials in Oncology Scholar Award (JSB); the Deutsche José Carreras Leukämie-Stiftung DJCLS 10R/2021 (TH); and by an allocation of computing resources from The Ohio Supercomputer Center and Shared Resources (Leukemia Tissue Bank). Support to Alliance for Clinical Trials in Oncology and Alliance Foundation Trials programs is listed at https://acknowledgments.alliancefound.org. Trial registration numbers of companion studies registered at www.clinicaltrials.gov are NCT00048958 (CALGB 8461), NCT00899223 (CALGB 9665) and NCT00900224 (CALGB 20202).
Author information
Authors and Affiliations
Contributions
Contribution: MO, A-KE, ASM, and KM designed the study; MO, DN, ASY, JK, KM, CJW, CCO, SO, JSB, J Buss, KTL, JCB, ASM, HLG, A-KE, NS, and VJ analyzed the data; DN and JK performed the statistical analysis; MO, KM, JK, DN, CJW, ASM, CCO, SO, JSB, JCB, and A-KE wrote the manuscript; WS, GLU, AJC, KM, WGB, BLP, JEK, JOM, RJM, RAL, JCB, TH, MR-T, AD, SS, MCS, DG, UK, WEB, BJW, WH, J Braess, MS, KS, and KHM were involved directly or indirectly in the care of patients and/or sample procurement; and all authors read and agreed upon the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
CJW is a consultant for Vigeo Therapeutics and employed at Karyopharm Therapeutics; and has ownership interest in Karyopharm Therapeutics and Bristol-Myers Squibb Co. JSB is a consultant/advisory board member for AbbVie, AstraZeneca, INNATE, KITE. BLP received honoraria from Jazz Pharmaceuticals, Novartis, and Pfizer. JEK has received honoraria from Gilead, Magellan, and Novartis; consulting fees from Gilead, Magellan, Novartis, Pharmacyclics, and Seattle Genetics; institutional research funding from Boehringer Ingelheim, Cantex, Erytech, and Millennium; and travel support from Gilead, Novartis, and Seattle Genetics. JCB has a consultancy/advisory role with Syndax, Novartis, Vincera; research funding from Pharmacyclics LLC, an AbbVie Company, Genentech, Janssen, Acerta; ownership for Vincera. A-KE has a current employment of her spouse at Karyopharm Therapeutics. The other authors declare no competing financial interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ozga, M., Nicolet, D., Mrózek, K. et al. Sex-associated differences in frequencies and prognostic impact of recurrent genetic alterations in adult acute myeloid leukemia (Alliance, AMLCG).
Leukemia (2023). https://doi.org/10.1038/s41375-023-02068-8
-
Received:
-
Revised:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41375-023-02068-8